DNA detectives to combat animal disease
When birds fall out of the sky, fish float lifeless in the river, or bats drop dead from trees, we scramble to find a cause. When mystery illnesses spread through our aquaculture fisheries or sheep flocks sicken, the race to identify a cause and a cure is even faster, given how quickly disease can spread in our intensive farming industries. Mr Sam Hair at the WA Department of Primary Industries and Regional Development (DPIRD) is using next-generation sequencing methods and Pawsey supercomputing for data analysis to quickly identify the pathogens causing mystery disease outbreaks, reducing the time to respond.
The Challenge
The Animal Pathology Laboratory at DPIRD works to protect the WA community and agriculture and food sector from the impacts of diseases and pests that affect animals and aquatics. When a mystery illness affects a herd or a sudden fish kill chokes a river, Mr Hair uses all of the tests at his disposal to identify the cause. But standard diagnostic tests are time-consuming, and sometimes fail.
“If the illness is caused by a brand-new strain of a virus, our standard diagnostic tests based on older strains may not identify it,” explains Mr Hair. “And when time is of the essence, testing for one potential cause after another and progressively ruling them out is time-consuming. Sometimes we can exhaust our battery of tests and still not find a cause.”
Next-generation sequencing raises the possibility of amplifying and expressing all of the genetic material found in an animal sample, including lurking bacteria and viruses. If the fragments can be stitched into complete genomes, these can be compared against international genome databases to identify the mystery pathogens that shouldn’t normally be there.
“The problem is that assembling all of the genetic material and comparing it to a database of millions of genetic sequences also takes time. Data analysis is the real bottleneck.”
The Solution
Mr Hair undertook a Pawsey Uptake project, working with supercomputing applications specialist Dr Marco De La Pierre to speed up the analytical work required to turn gene fragments into a disease diagnosis.
“We still do the genome sequencing in the DPIRD laboratory, but instead of processing those fragments on our local desktop computers, we port the data over to the Pawsey systems. In the Uptake Project we created an automated workflow that assembles the genomes and then compares them against existing databases.”
The workflow reduces the effort Mr Hair spends on repetitive analysis, genome assembly, quality control and database searching to just a few computer commands to analyse a collection of samples. The result is either a match for a known disease pathogen, or the complete genome of something new, which can become the basis for a more targeted and routine diagnostic test.
The Outcome
“If an animal is sick or dead from something completely unknown, we need a diagnostic protocol that covers all of the possibilities and identifies everything there, not just a test that may confirm what we might suspect. Next-generation sequencing gives us that capability,” points out Mr Hair. “But it is only useful if we can analyse enough samples and process the data fast enough to make a difference to a real-world scenario.”
Investigating a recent fish-kill incident highlights the power of the DNA detective approach. Whereas Mr Hair would have previously tested for likely disease culprits for weeks until he got a positive result, next-generation sequencing was used to very quickly rule out the involvement of any pathogen at all, allowing the Department to turn its focus immediately to environmental factors instead.
Dr De La Pierre is continuing to work with Mr Hair to streamline the workflow, improve its speed and ease of use, and enable data from multiple genetic sequencing technology platforms to be used. The workflow is available to any research organisation accessing Pawsey facilities, so it can be used by other researchers to quickly assemble and identify any genomes of interest, regardless of where the sample came from.
But for Mr Hair and DPIRD, it’s Western Australia’s newest tool in bio-surveillance and disease investigation to support animal welfare, food safety and agricultural production.
Project Leader.
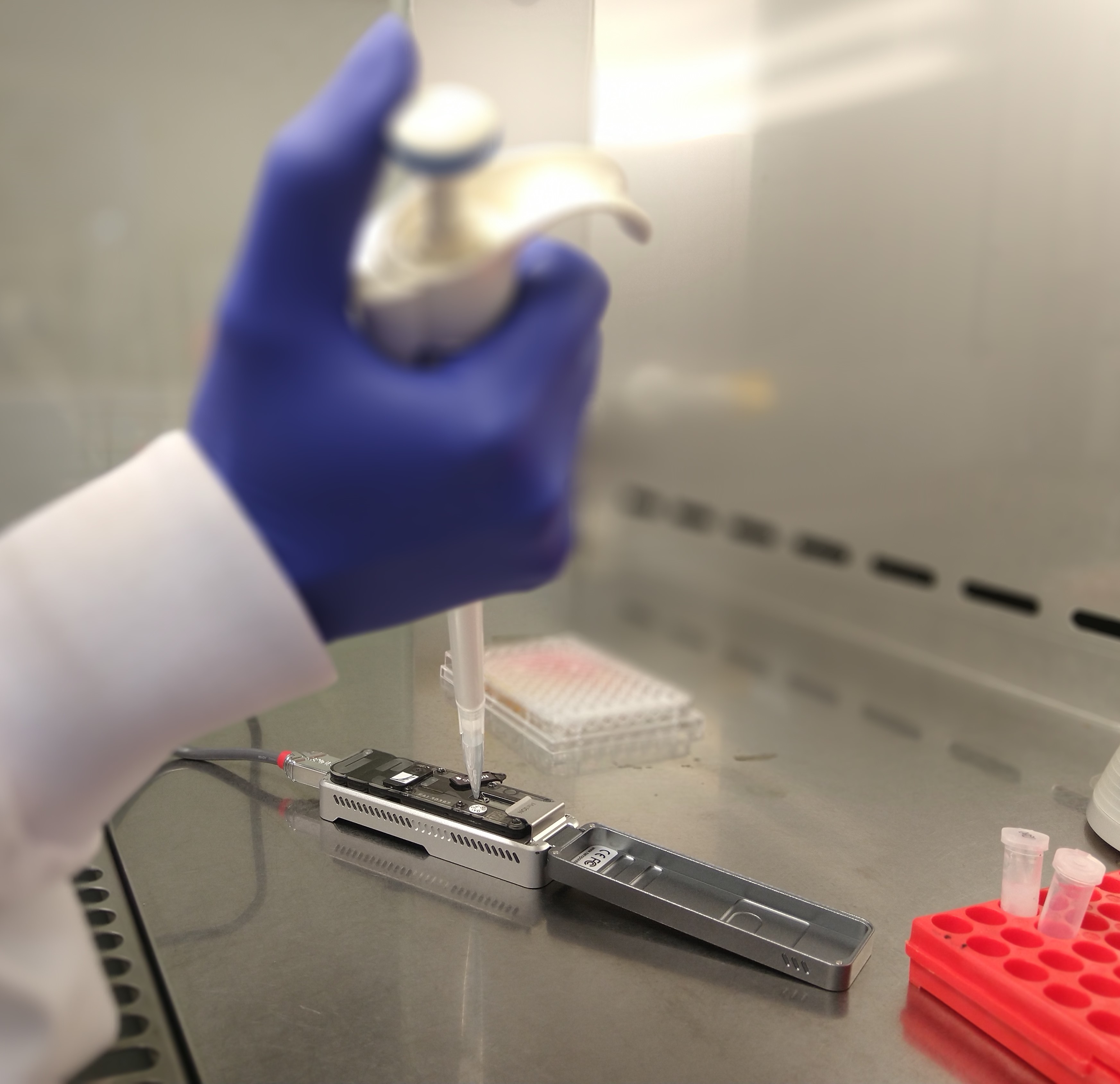
Image courtesy of Mr Sam Hair, WA Department of Primary Industries and Regional Development (DPIRD)

Sheep at Murdoch University. Image courtesy of Mr Sam Hair, WA Department of Primary Industries and Regional Development (DPIRD)