Shared workflows, virus’ lineage, statistical inferences about potential treatment outcomes, COVID-19 targets molecular modelling are some of the collaborations taking place at the two national tier-1 supercomputing facilities to fight the pandemic.
In early April, Australia’s Tier 1 supercomputing facilities, Pawsey Supercomputing Centre (Pawsey) and National Computational Infrastructure (NCI), joined forces to offer additional computation and data resources to support the national and international research community to acquire, process, analyse, store and share data supporting COVID-19 research.
Both facilities contributed extensive resources to assist researchers in the fight to overcome COVID-19. Through this initiative, researchers throughout Australia are now working with NCI which is currently supporting three targeted projects with more than 40 million units of compute time on the Gadi supercomputer; and Pawsey Supercomputer Centre, which provides access across five projects to over 1100 cores on the newly deployed Nimbus cloud and Topaz.
On July 31st, five researchers came together on Zoom to show over 70 attendees how they are using these facilities to understand the lineage of the SARS-COV-2 coronavirus while making statistical inferences about potential treatment outcomes, molecular modelling and more.
When asked how critical the supercomputing infrastructure is to their research, parallels were drawn between standard desktops and supercomputers. If this COVID-19 research was undertaken on a standard computer, it would take at least 26 years to achieve the work that will be completed by March 2021, using supercomputers. This is their work:
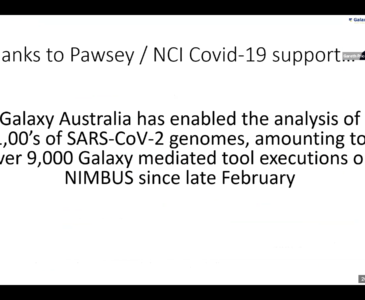
Dr Gareth Price, Head of Computational Biology at QCIF Facility for Advanced Bioinformatics showcased how Galaxy Australia has a dedicated COVID-19 pulsar, which is hosted and supported by the Pawsey Nimbus cloud service. The COVID-19 pulsar, which is accessible across the global Galaxy network, is a resource is to provide publicly accessible infrastructure and workflows for SARS-CoV-2 data analyses. Currently, they have three different types of analyses openly available – genomics, evolution and cheminformatics. These analyses and their best practices are available at https://covid19.galaxyproject.org/.
“Galaxy Australia, an open-sourced platform available for computational biological research across the world, has enabled the analysis of hundreds of SARS-CoV-2 genomes, amounting to over 9000 Galaxy mediated tool executions on Nimbus since late February,” said Dr Price during his presentation.
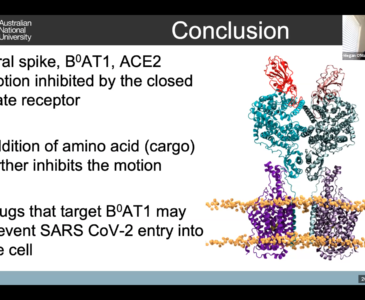
A/Prof Megan O’Mara at Australian National University showed viewers how her team uses large-scale molecular dynamics for rational drug design. Using the Gadi supercomputer at NCI, Megan’s team uses simulations of approximately 800,000 atoms that make up a key receptor of the human body to understand exactly how the coronavirus uses it to invade human cells. It is only with high-resolution modelling, achieved by supercomputers, can they accurately replicate the true behaviours of the receptors and figure out where vulnerabilities in the virus’ binding process are.
This means that in the future, this understanding of receptors will assist in the development of drug targeting to prevent viruses such as SARS-CoV-2 from entering the human systems.
Supercomputing has not only allowed the team to target interactions to develop a drug design against COVID-19, but it has allowed them to pursue a holistic understanding of the virus, to anticipate and protect us from potential threats in the future.
Dr Tom Karagiannis at Monash University together with Dr Andrew Hung from RMIT University also uses molecular modelling for COVID-19 targets. In the absence of a vaccine for the virus, Tom’s group repurposes existing research for potential antiviral effects. His team used GPU resources on Topaz to provide a molecular basis for known antivirals that could potentially stop the virus from replication inside our cells. and identify new ones which offer a protective effect.
Tom’s team has been using supercomputer to model the behaviour of a molecule called a-ketoamide. It was chosen because it is an antiviral with a broad spectrum developed by a German Lab for SARS-1. Based on his findings, the α-ketoamide compound class could be pursued as potential antivirals against COVID-19. You can read more in his recent publication here.
He is currently researching a possible target site on the SARS-CoV-2 main protease dimer interface.
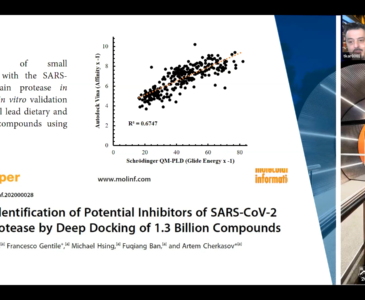
With this information, Tom’s group is now focused on high-throughput screening of a library of small molecules to identify additional lead compounds with potential antiviral activity. The GPU resources on Topaz enabled the group to investigate more realistic binding of the small molecules to the main protease, allowing for selection of a final group of compounds for validation in the laboratory.
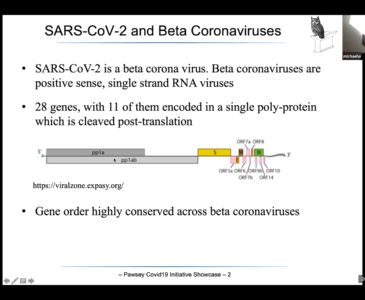
A/Prof Michael Wise at the University of Western Australia is a computational biologist who has a history of researching endemics, such as SARS, MERS and now SARS-CoV-2 (the COVID-19 coronavirus). His years of research works on the basis that these viruses all share a core proteome (the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time). Michael’s team reconstructs the phylogenetic trees in which SARS and SARS-CoV-2 are placed. An understanding of this and its core-proteome means that drugs and antivirals can be targeted at their core as opposed to their strains; protecting us in the future from new viruses.
Prof Wise has recently preprint of the first results on the OSF platform.
Shelley Barfoot at the University of Queensland is part of a team of researchers led by Dr Alan Mark. Their team is understanding and modelling the key structural changes in the protein that allows a coronavirus to enter and infect a human cell. This project aims to throw light on a critical target for both vaccine development and the discovery of antiviral agents. In particular, working with researchers from one of three centres worldwide charged with rapid vaccine development by the World Health Organisation, the aim is to understand how potential vaccine constructs that incorporate the coronavirus fusion protein can be optimised.
This team uses Australia’s national supercomputing facilities to assist in the global effort to identify existing drugs that could be repurposed to treat COVID-19. The Automated Topology Builder (atb.uq.edu.au), a globally recognised molecular modelling tool developed at The University of Queensland, will be used to develop high-quality atomic interaction parameters for all pharmaceutically active compounds that have passed phase 2 clinical trials (efficacy and safety). Read more about the ATB and its impact here.
Missed out?
Did you miss this Pawsey Friday? It is now available on Pawsey’s YouTube channel to watch.
Want to be notified of upcoming events and training? Sign up to be a Pawsey Friend so you don’t miss out again.

Dr Gareth Price, Head of Computational Biology at QCIF Facility for Advanced Bioinformatics
A/Prof Megan O’Mara at Australian National University
Dr Tom Karagiannis at Monash University
A/Prof Michael Wise at the University of Western Australia
Shelley Barfoot at the University of Queensland

